Eukaryotic transcription factors can track and control their target genes using DNA antennas
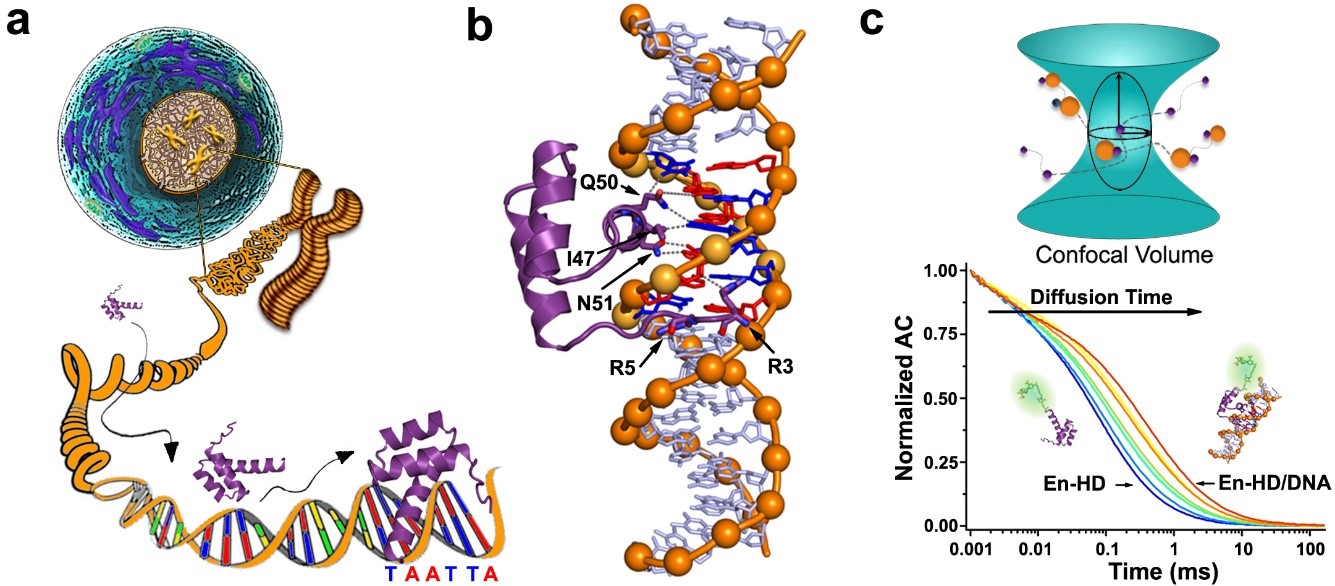
Engrailed Homeodomain (EngHD) binding to the target specific site. a) Pictorial representation of the challenges involved in tracking the target cognate site in the genomic DNA of a eukaryotic cell. b) 3D Structure of the specific complex between EngHD and DNA (PDB: 1HDD). c) Schematic diagram of how to determine binding of EngHD to DNA using FCS.
05.02.2020
Eukaryotic transcription factors (TF) function by binding to short 6-10 bp DNA recognition sites located near their target genes, which are scattered through vast genomes. Such process surmounts enormous specificity, efficiency and celerity challenges using a molecular mechanism that remains poorly understood. Combining biophysical experiments, theory and bioinformatics, the authors dissect the interplay between the DNA-binding domain of Engrailed, a Drosophila TF, and the regulatory regions of its target genes. They find that Engrailed binding affinity is strongly amplified by the DNA regions flanking the recognition site, which contain long tracts of degenerate recognition-site repeats. Such DNA organization operates as an antenna that attracts TF molecules in a promiscuous exchange among myriads of intermediate affinity binding sites. The antenna ensures a local TF supply, enables gene tracking and fine control of the target site’s basal occupancy. This mechanism illuminates puzzling gene expression data and suggests novel engineering strategies to control gene expression.
A research team at IMDEA NANOCIENCIA, guided by Prof. Víctor Muñoz (current position at UCMerced: https://www.ucmerced.edu/), has addressed a fundamental question in eukaryotic genes regulation, and for that they utilize biophysical methods to dissect the binding process, statistical mechanical modeling to integrate and rationalize the results, and bioinformatics analysis to further explore the functional implications. Eukaryotic TFs track their target genes, control site occupancy, and coordinate binding with partners to form the transcription complex. These processes must involve modes of interaction with DNA that go beyond nonspecific binding and facilitated 1D and/ or 2D diffusion. By focusing on the DNA regions flanking the target site of real genes, they have discovered, and characterized, a molecular mechanism that enables such functions. The mechanism exploits the natural tendency of biomolecules to exhibit energetic frustration, in this case manifested by binding promiscuity. Particularly, they find that the affinity of the Drosophila TF Engrailed to the regulatory regions of its target genes is strongly amplified by long tracts of degenerate consensus repeats that are present in such regions. The combination of a promiscuous TF and a DNA region rich in degenerated consensus binding repeats operates as a transcription antenna. The present research provides an elegant mechanism that sheds new light on how eukaryotic TFs operate at the molecular level and explains several paradoxes of existing eukaryotic gene expression data. These molecular devices provide an additional layer of eukaryotic transcriptional control in which the size and sequence profile of the antenna can be engineered, whether by evolution or by scientists, to modulate site occupancy, response swiftness and levels of gene expression.
This work was supported by the European Research Council through grant ERC-2012-ADG-323059. M.C. acknowledges support from a Juan de La Cierva fellowship from the Spanish Ministry of Economy and Competitiveness. V.M. acknowledges support from the W.M. Keck foundation and the National Science Foundation through grants NSFCREST-1547848 (Center for Cellular and Biomolecular Machines) and NSF-MCB1616759.
Keywords: Transcription Factors, Eukaryotic gene expression regulation, Biophysics of DNA-Protein interactions, FCS, Engrailed.
Reference:
M. Castellanos et al. Nat. Comm. (2020)
https://doi.org/10.1038/s41467-019-14217-8
Contact:
Dr. Milagros Castellanos
This email address is being protected from spambots. You need JavaScript enabled to view it.
Prof. Víctor Muñoz
This email address is being protected from spambots. You need JavaScript enabled to view it.
IMDEA Nanociencia Outreach Office
This email address is being protected from spambots. You need JavaScript enabled to view it.
+34 912998712



